Current Highlights
Here’s a selection of projects that I’ve worked on. For each project, I’ve included a brief description, some of the technologies used, and links to view more details or the source code.
Gaussian Processes and Multi-Objective Bayesian Optimization
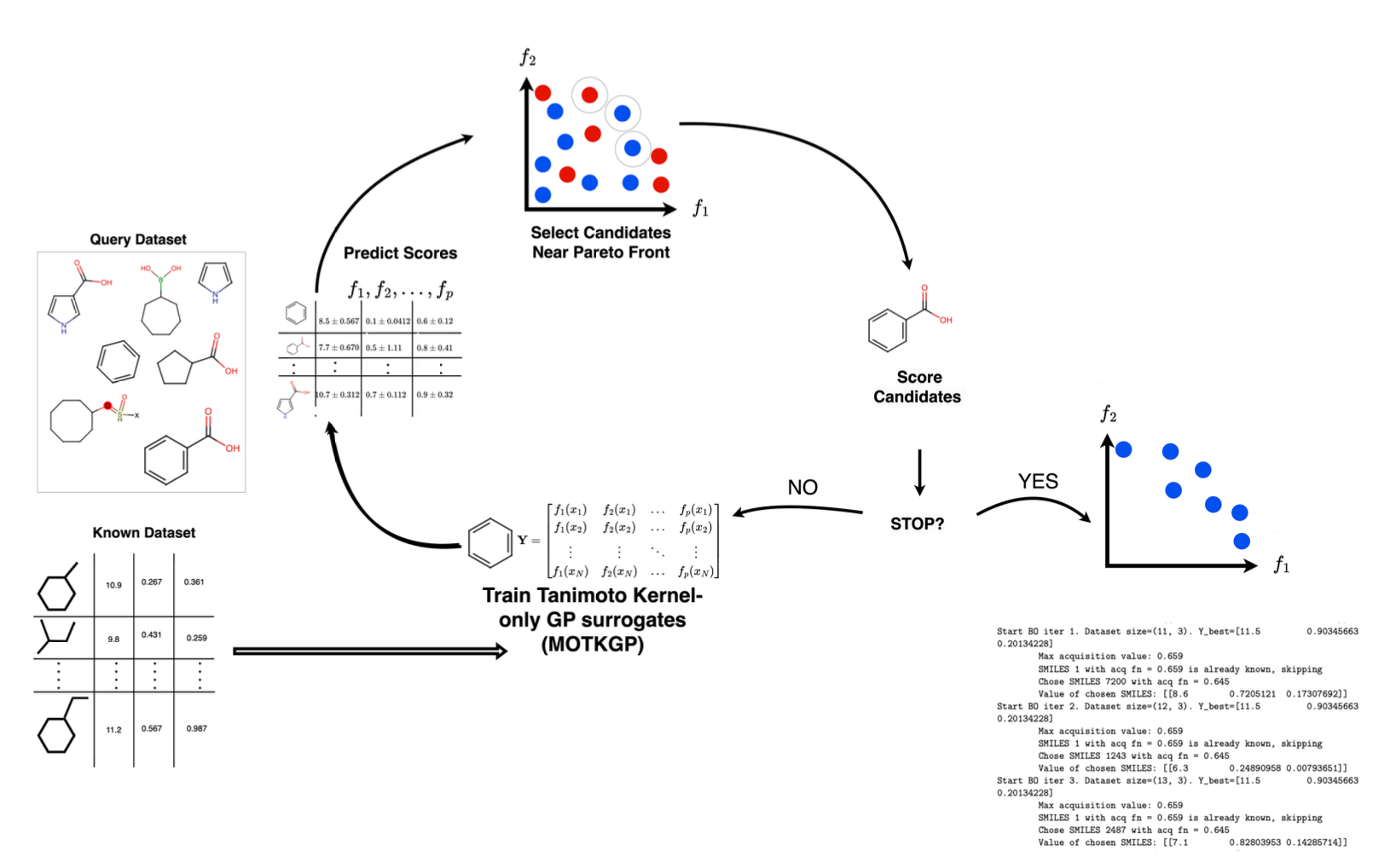 Description: For my masters thesis, we present GP-MOBO, a novel multi-objective Bayesian Optimization algorithm that
advances the state-of-the-art in molecular optimization. Our approach integrates a fast minimal package for Exact Gaussian Processes (GPs) capable of efficiently handling the full dimensionality of sparse molecular fingerprints without the need for extensive computational resources.
Description: For my masters thesis, we present GP-MOBO, a novel multi-objective Bayesian Optimization algorithm that
advances the state-of-the-art in molecular optimization. Our approach integrates a fast minimal package for Exact Gaussian Processes (GPs) capable of efficiently handling the full dimensionality of sparse molecular fingerprints without the need for extensive computational resources.
- Technologies used: Python
- Skills attained: Multi-Objective Bayesian Optimization, Kernel Methods, Gaussian Processes, Molecular Optimization with Generative Models
- View Project
- Source Code
Probabilistic and Unsupervised Learning
 Description: Arguably the most interesting and difficult machine learning I have done! This spans from probabilistic modelling with multivariate Gaussians, model selection, expectation maximisation algorithm with binary data, linear gaussian state space models (LGSSMs), Monte Carlo Markov Chain methods (MCMC) for message decryption, Gibbs sampling with latent distributed allocation, and optimization problems. A course requirement for Gatsby Computational Neuroscience Unit PHD module
Description: Arguably the most interesting and difficult machine learning I have done! This spans from probabilistic modelling with multivariate Gaussians, model selection, expectation maximisation algorithm with binary data, linear gaussian state space models (LGSSMs), Monte Carlo Markov Chain methods (MCMC) for message decryption, Gibbs sampling with latent distributed allocation, and optimization problems. A course requirement for Gatsby Computational Neuroscience Unit PHD module
- Technologies used: Python, MATLAB, Stata
- Skills attained: Unsupervised Learning, Bayesian Statistics, Graphical Models (Markov networks and Bayesian networks)
- View Project
- Source Code
Statistical Natural Language Processing
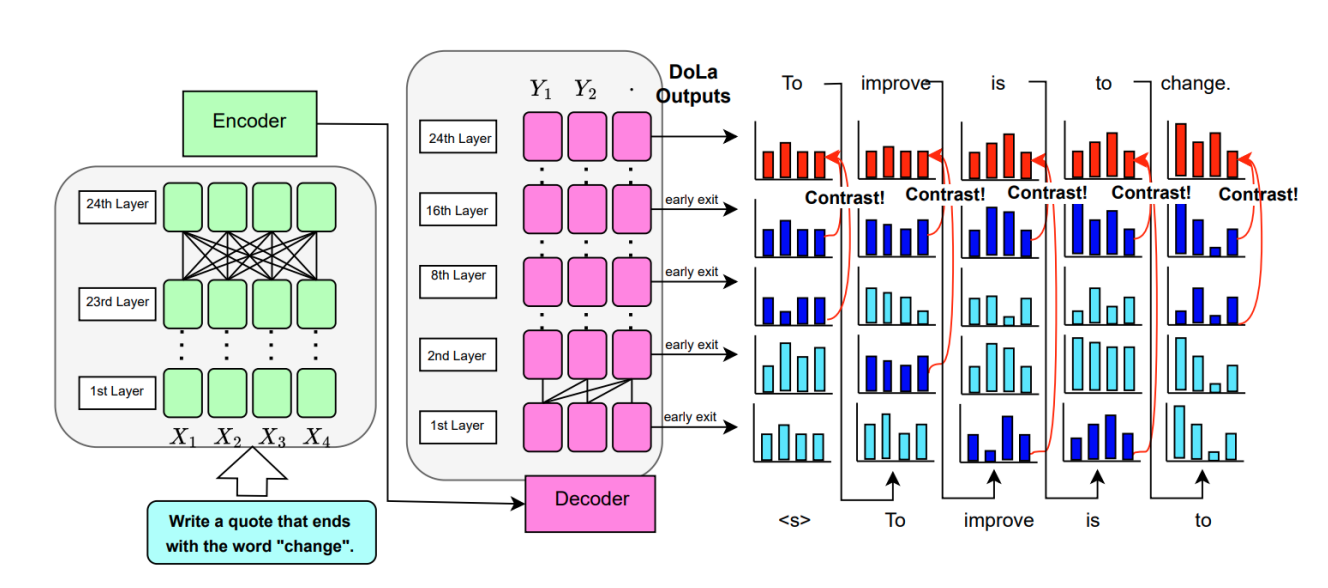 Description: This project adapts the DoLa contrastive decoding method to encoder-decoder models like T5 and FLAN-T5 to improve instruction-following performance. While DoLa improves faithfulness in certain tasks (e.g., keyword inclusion), it harms others, highlighting its context-dependent effectiveness.
Description: This project adapts the DoLa contrastive decoding method to encoder-decoder models like T5 and FLAN-T5 to improve instruction-following performance. While DoLa improves faithfulness in certain tasks (e.g., keyword inclusion), it harms others, highlighting its context-dependent effectiveness.
- Technologies used: Python
- Skills attained: LLM hallucinations, Contrastive Decoding strategies for LLMs
- View Project
- Source Code
Numerical Optimization: Developing A New Subspace Newton Method!
 Description: This project implements a Subspace Newton Method for Sparse SVMs (NSSVM) to solve large-scale binary classification problems efficiently using a sparsity-constrained kernel-based optimization. The method achieves fast convergence, reduced model complexity, and competitive accuracy by adaptively tuning the number of support vectors and exploiting strong convexity in the dual formulation.
Description: This project implements a Subspace Newton Method for Sparse SVMs (NSSVM) to solve large-scale binary classification problems efficiently using a sparsity-constrained kernel-based optimization. The method achieves fast convergence, reduced model complexity, and competitive accuracy by adaptively tuning the number of support vectors and exploiting strong convexity in the dual formulation.
- Technologies used: Python
- Skills attained: Sparsity-Constrained Optimization, Subspace Newton Method, Quasi-Newton methods (e.g., BFGS, L-BFGS), Lipschitz continuity
- View Project
- Source Code
My Past Projects
Bayesian MCMC for ABO Blood Frequency Modelling
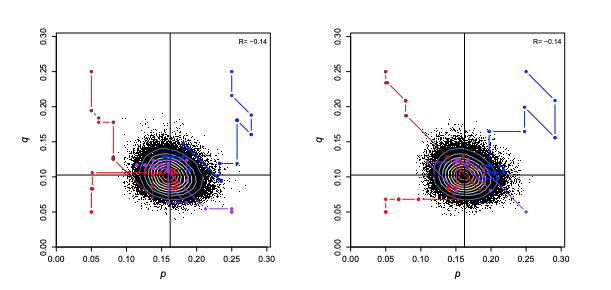 Description: Estimation of allele frequencies in the ABO blood group, The ABO blood type is determined by the presence or absence of the A and B antigens on erythrocytes. It is controlled by a single gene (the ABO gene) with three alleles: I^A, I^B, and i. Here I stands for isoagglutinogen or antigen, while i means absence of either antigen. For convenience we write the three alleles as A,B, and O. As both A and B alleles are dominant over O, genotypes AA or AO both have the same phenotype (type A), and individuals with BB
or BO have type B. At Hardy-Weinberg equilibrium, the genotype and phenotype frequencies are given as functions of the frequencies of the three alleles, p, q, and r = 1− p−q. The data, X = (nA,nB,nAB,nO), are counts of the four blood types.
Description: Estimation of allele frequencies in the ABO blood group, The ABO blood type is determined by the presence or absence of the A and B antigens on erythrocytes. It is controlled by a single gene (the ABO gene) with three alleles: I^A, I^B, and i. Here I stands for isoagglutinogen or antigen, while i means absence of either antigen. For convenience we write the three alleles as A,B, and O. As both A and B alleles are dominant over O, genotypes AA or AO both have the same phenotype (type A), and individuals with BB
or BO have type B. At Hardy-Weinberg equilibrium, the genotype and phenotype frequencies are given as functions of the frequencies of the three alleles, p, q, and r = 1− p−q. The data, X = (nA,nB,nAB,nO), are counts of the four blood types.
- Technologies used: Python, R
- Skills attained: Statistical Analysis, Hypothesis Testing, Bayesian Modelling
- View Project
- View Project 2
- Source Code
Using Structural Bioinformatics Approach for GULO functionality
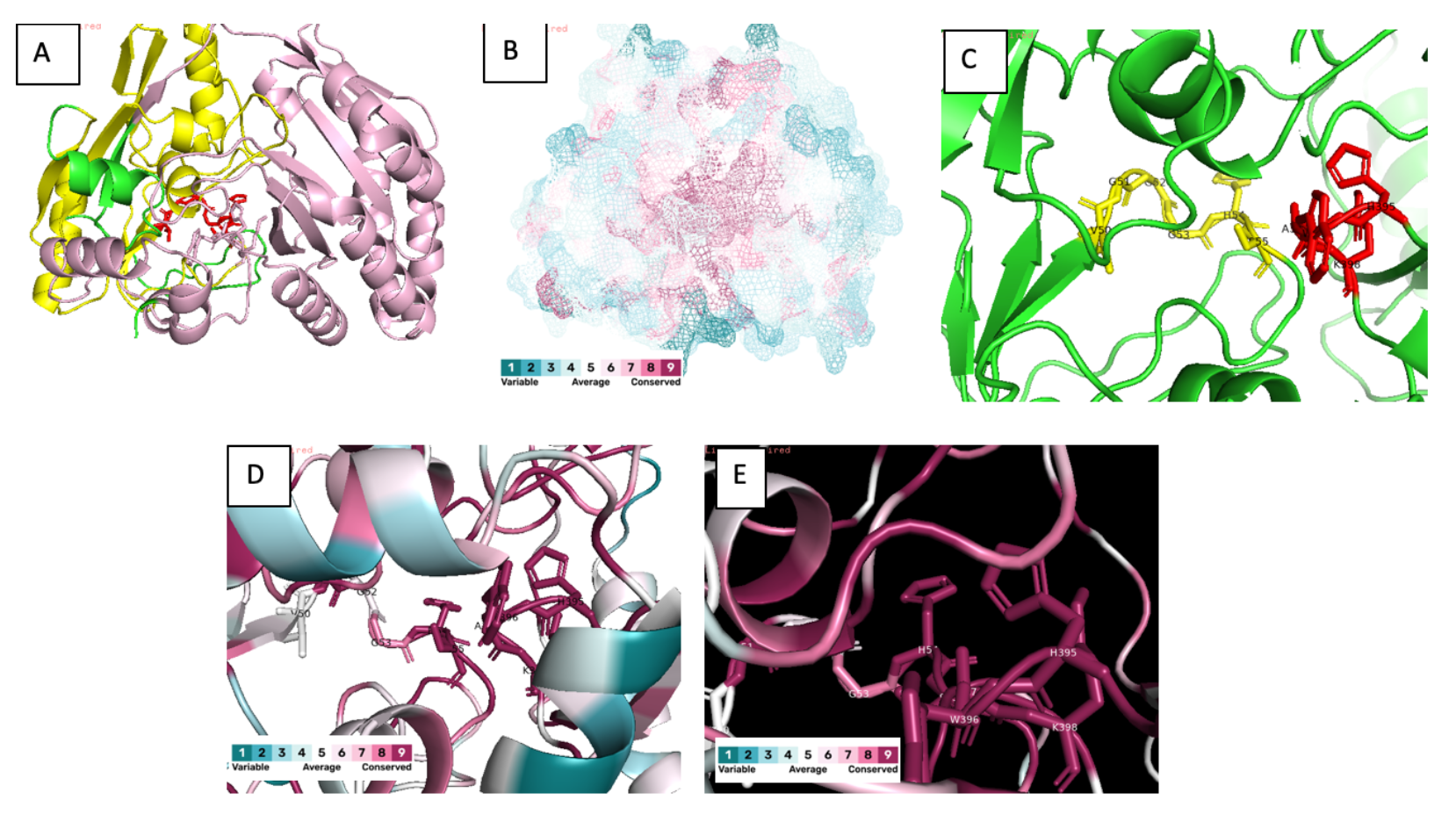 Description: Vitamin C is an important cofactor in many important physiological processes. Vitamin C deficiency usually leads to diseases such as scurvy.The GULO gene encodes an enzyme which converts L-gulono-1,4-lactone to L-ascorbate (vitamin C). GULO enzyme (L-gulonolactone oxidase) is required in the terminal step of catalytic reaction. Members of this enzyme fam- ily contain two important domains: FAD-binding domain and ALO domain. Here demonstrates a structural bioinformatics and functional genomics approach to determine the functionality of GULO gene in certain species.
Description: Vitamin C is an important cofactor in many important physiological processes. Vitamin C deficiency usually leads to diseases such as scurvy.The GULO gene encodes an enzyme which converts L-gulono-1,4-lactone to L-ascorbate (vitamin C). GULO enzyme (L-gulonolactone oxidase) is required in the terminal step of catalytic reaction. Members of this enzyme fam- ily contain two important domains: FAD-binding domain and ALO domain. Here demonstrates a structural bioinformatics and functional genomics approach to determine the functionality of GULO gene in certain species.
- Bioinformatics Software & Databases Utilized: PYMOL, CONSURF, UniProt, ProteinDataBank(PDB), GenBank, NCBI Blastn, Blastp, tBlastn
- Skills attained: Statistical Analysis, Hypothesis Testing, Biochemistry, Gene Ontology
- View Project
Biochemistry-focused Projects
Investigating molecular mechanism behind DNMT1 methylation of CpG islands
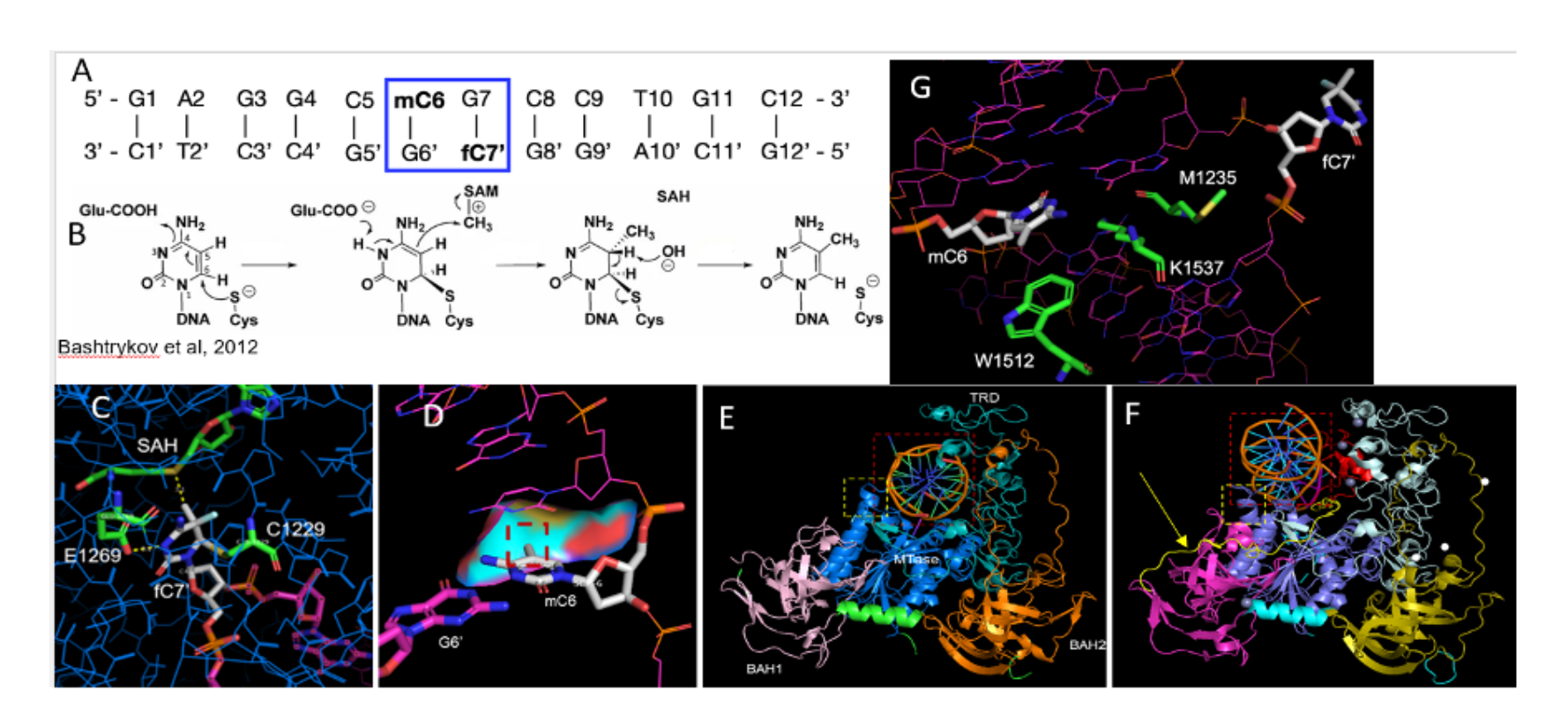 Description: I investigated the molecular mechanism of mouse DNMT1-DNA structures with available PDB structures DNMT1 bound to hemi-methylated DNA (4DA4) and unmethylated DNA (3PT6) found on UniProt and ProteinDataBank.
Description: I investigated the molecular mechanism of mouse DNMT1-DNA structures with available PDB structures DNMT1 bound to hemi-methylated DNA (4DA4) and unmethylated DNA (3PT6) found on UniProt and ProteinDataBank.
- Bioinformatics Software & Databases Utilized: PYMOL, ClinVar, UK BioBank. CONSURF, UniProt, ProteinDataBank(PDB)
- View Project
Investigation of molecular mechanism of Minichromosome Maintenance Protein (MCM) protein-DNA interactions
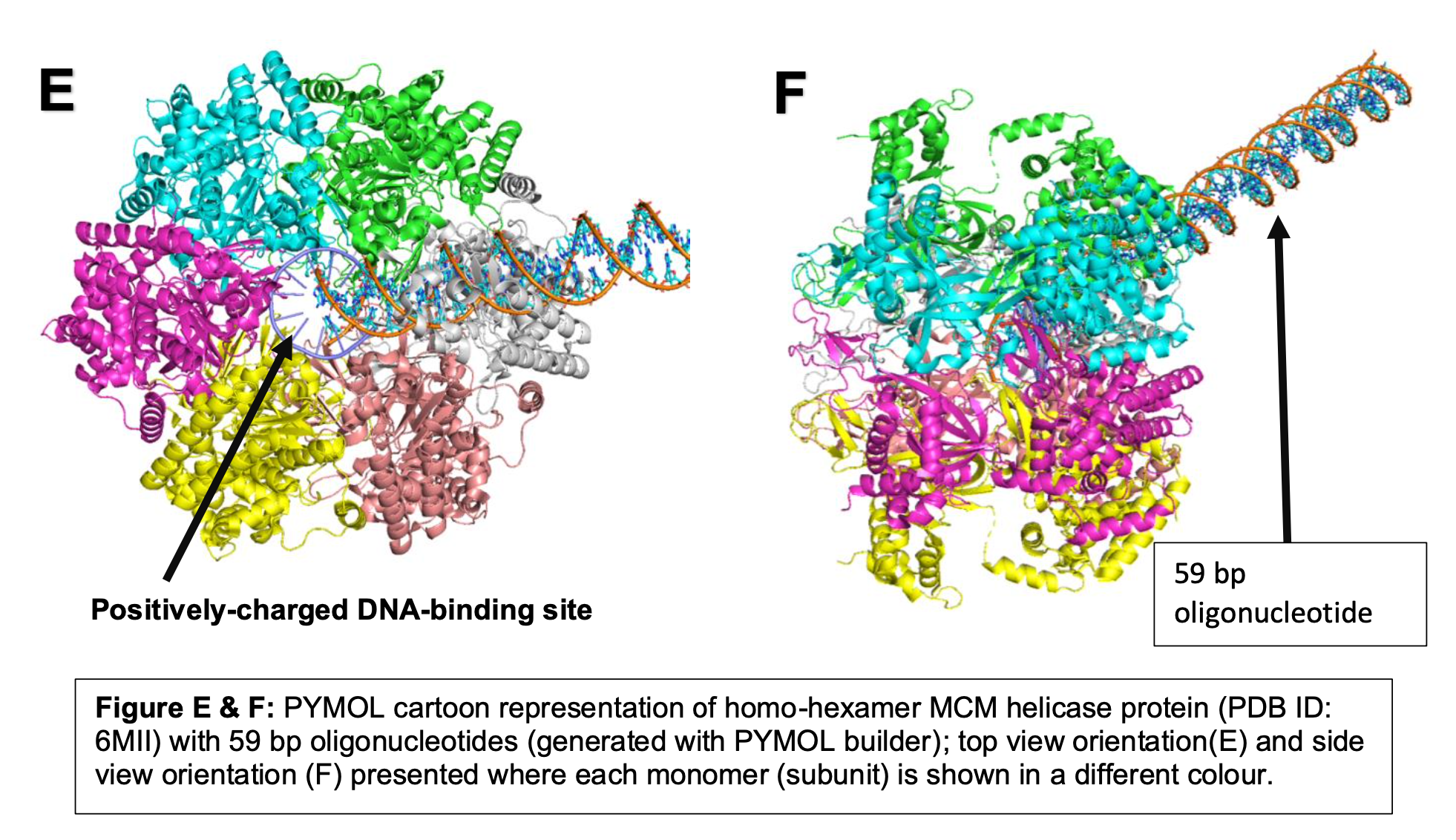 Description: Part of my biochemistry module, we utilized both wet lab and bioinformatics approaches to investigate the molecular mechanism of MCM protein.
Description: Part of my biochemistry module, we utilized both wet lab and bioinformatics approaches to investigate the molecular mechanism of MCM protein.
- Bioinformatics Software & Databases Utilized: PYMOL, CONSURF, UniProt, ProteinDataBank(PDB)
- Skills attained: Biochemistry, SDS-Page Gel Electrophoresis, Electrophoretic Mobility Shift Assay, Bioinformatics
- View Project
For inquiries or further information about my work, feel free to email me or check out my GitHub profile.