Research Experiences
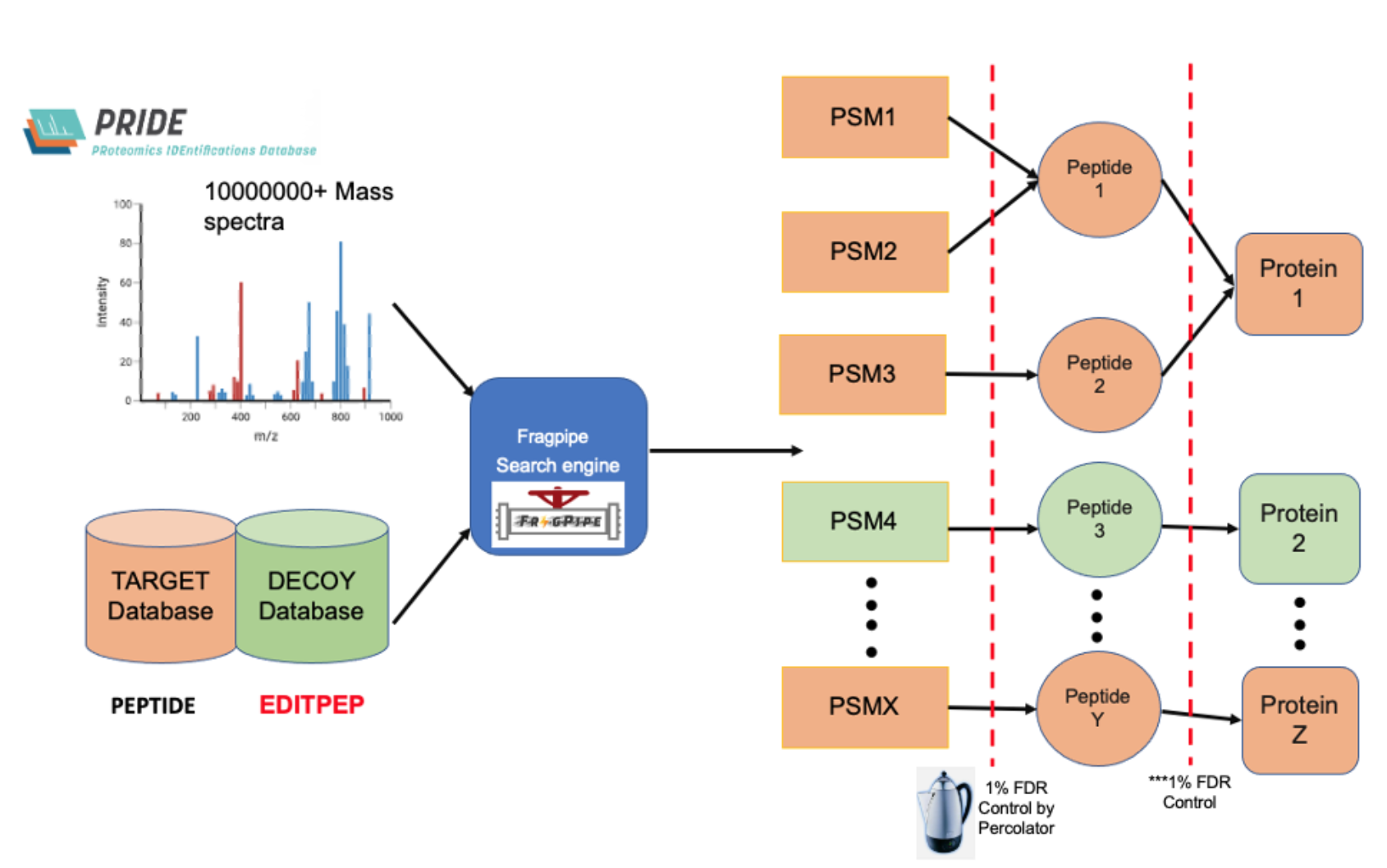 Computational Biologist @ University of Edinburgh, under supervision of Professor Georg Kustatscher
Computational Biologist @ University of Edinburgh, under supervision of Professor Georg Kustatscher
- Optimised data preprocessing tool for combining multiple large-scale MaxQuant searches on protein group-level pgFDR, which let to discovering 7000+ novel microproteins
- Implemented this tool for accurate & sensitive protein group FDR method on MaxQuant and Fragpipe database outputs with protein isoforms on ProteomeHD2 dataset curated by Professor Georg Kustatscher
- Controlled false discovery rate for feature selection in human MS-based proteomics dataset (size: 27.1 Gb)
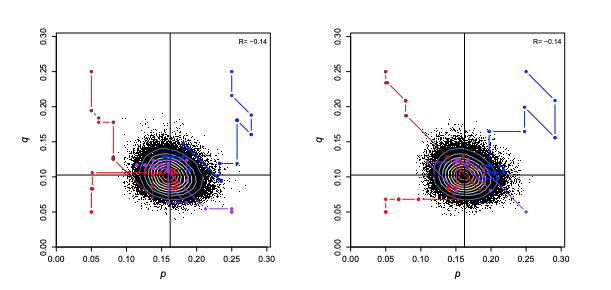 Statistical Geneticist @ UCL Centre of Computational Biology, under supervision of Professor ZiHeng Yang and Tomas Fluori
Statistical Geneticist @ UCL Centre of Computational Biology, under supervision of Professor ZiHeng Yang and Tomas Fluori
- Developed statistical models for predictive phylogenetic analysis on blood allele frequencies provided in a small dataset.
- Implemented Bayesian MCMC algorithm to sample probability distributions in p, q, and r frequencies from Hardy-Weinberg principle.
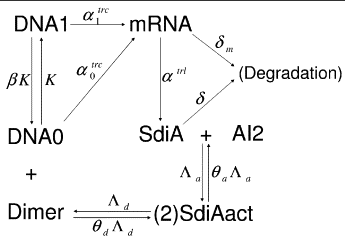 Mathematical Biologist @ Centre for Systems Biology Edinburgh, under supervision of Professor Ramon Grima
Mathematical Biologist @ Centre for Systems Biology Edinburgh, under supervision of Professor Ramon Grima
- Developed stochastic models (stochastic simulation algorithm) & deterministic models to analyse and predict complex data patterns in mRNA decay biochemical kinetics.
- Conducted extensive research in Continuous Time Markov Chain processes for sampling functional Kolmogorov Forward Equations (chemical Master Equation).
- Applied Gibbs sampling (MCMC) and Metropolis Hastings algorithm to investigate consistent genes in the bipartite network
Previous work (View Projects for Softwares and Tools Utilized)
-
Evaluation of different False discovery rate approaches in Large-scale Proteomics Data
Bachelor’s Thesis, supervised by Georg Kustatscher.
Achieved an 80% (A2).
View PDF → -
Random Forest with Bayesian Optimisation for Heart Failure Prediction
View PDF → -
Using structural bioinformatics approach for GULO functionality
Achieved a 95% (A1).
View PDF → -
Purification and Quantification of protein-DNA interactions of Minichromosome Maintenance Protein (MCM) of Pyrococcus abyssi
Achieved a 90% (A1).
View PDF → -
DNMT1 methylates hemi-methylated CpGs but not un- methylated CpGs View PDF →